| Visit DIX: German-Spanish-German dictionary | diccionario Alemán-Castellano-Alemán | Spanisch-Deutsch-Spanisch Wörterbuch |
Functions in this class mainly provide a way to simple use tensors and
apply basic mathematical operations on them.
The data stored in a tensor is a floating point precision array:
float _t[9]
where the indexing is
![\begin{displaymath}
\mathtt{
\left(\begin{array}{ccc}
\_t[0] &\_t[1] &\_t[2]\\
...
...t[4] &\_t[5]\\
\_t[6] &\_t[7] &\_t[8]\\
\end{array}\right)
}
\end{displaymath}](img383.gif)
The only private function is
void eigsort(const double *W, int *i) const;
which sorts the eigenvalues by size and is used in eig(...).
Three different constructors allow the generation of an empty
tensor (all values are set to zero), a symmetric tensor where the
indexing corresponds to
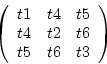
tensor();
tensor(float t1, float t2, float t3, float t4, float t5, float
t6);
tensor(float t1, float t2, float t3, float t4, float t5, float t6, float t7, float t8, float t9);
Three corresponding functions are available to set the values after the initialization:
void setZero();
void set(float t1, float t2, float t3, float t4, float t5,
float t6);
void set(float t1, float t2, float t3, float t4, float t5,
float t6, float t7, float t8, float t9);
float get(const int i, const int j) const;
returns the value at position ![]() in the tensor where
in the tensor where
![]() and
and
bool isZero() const;
checks if the tensor is empty, i.e. all values are zero and returns
true if so.
The basic mathematical operations can be performed between tensors and between tensors and scalars. Dividing a scalar by a tensor is not a legal operation.
tensor operator+ (const tensor &s, const tensor &t);
tensor operator+ (const tensor &t, const float x);
tensor operator+ (const float x, const tensor &t);
tensor operator- (const tensor &s, const tensor &t);
tensor operator- (const tensor &t, const float x);
tensor operator- (const float x, const tensor &t);
tensor operator* (const tensor &s, const tensor &t);
tensor operator* (const tensor &t, const float x);
tensor operator* (const float x, const tensor &t);
tensor operator/ (const tensor &s, const tensor &t);
tensor operator/ (const tensor &t, const float x);
tensor operator- (const tensor &t);
The last functions changes the sign of a tensor.
To send a tensor to the standard output the operator
ostream &operator « (ostream &out, const tensor &t);
is used, which will print the tensor values like
Tensor values:
![\begin{displaymath}
\begin{array}{lll}
[ \mathtt{5 } & \mathtt{ 20 } & \mathtt{...
...;}\\
\mathtt{ 25} & \mathtt{ 30 } & \mathtt{ 15 ]}\end{array}\end{displaymath}](img387.gif)
float det() const the determinant
float trace() const the trace (see invariantinequation 4.6)
tensor inv(tensor &result) const the inverse of a 33 matrix
tensor trans(tensor &result) const the transpose of a 33 matrix
To compute the eigenvalue, -vector decomposition (see equation 4.5) the CLAPACK function
dsygv_(...) is called. The eigenvalues are returned so that
they can be directly used to instantiate an eigensystem. The last
argument must be 1 if the eigenvalues and eigenvectors should be
sorted in descending order.
int eig( float &eig1, float &eig2, float &eig3, float *vector1, float
*vector2, float *vector3, const int sort=0) const;
The single value decomposition also calls a CLAPACK function (see
equation 7.30) whereas CLAPACK returns directly ![]() instead of
instead of ![]() .
.
int svd( float &r1, float &r2, float &r3, tensor &q, tensor &s);
The equivalence between equation 7.30 and the function call
is:
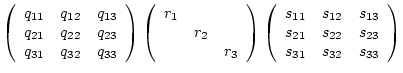 |
The following measurements can be computed:
float euclid() const euclidian magnitude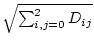
float Max() const maximal value in tensor
float Min() const minimum value in tensor
The boolean comparison between two tensors is based on the function
float measure()const {return euclid();};
so that ![]() ,
, ![]() ,
, ![]() and
and ![]() are defined. The comparisons
are defined. The comparisons ![]()
![]() and
and ![]() are based on every single component of the tensor.
are based on every single component of the tensor.
Use of the following functions should be avoided whenever possible
since they use the expensive eig(...) functions to compute the
eigenvalue, -vector decomposition and only return one value. It is
probably better to create an eigensystem (instance of class
eigen.h) and derive this measurements from there.
float* eigvec1(float *vector) const; first eigenvector, corresponding to largest eigenvalue
float* eigvec2(float *vector) const; second eigenvector
float* eigvec3(float *vector) const; third eigenvector, corresponding to smallest eigenvalue
float eigval1() const; first (largest) eigenvalue
float eigval2() const; second eigenvalue
float eigval3() const; third eigenvalue
float anisotropy() const; anisotropy, see equation 4.17
eigen(const float eig1, const float eig2, const float eig3, const float *vec1,To set values in an eigensystem there are several specialized functions:
const float *vec2, const float *vec3);
a constructor which initializes all values
eigen(); a constructor which sets all values to zero.
void set(const float eig1, const float eig2, const float eig3,const float *vec1,If the largest eigenvalue is zero then the whole eigensystem is zero. Therefore the last function only needs to set _lambda1 to zero.
const float *vec2, const float *vec3); set all the values in an eigensystem.
void set(const float eig1, const float eig2, const floateig3);
set the eigenvalues.
void setVectors(const float *vec1, const float *vec2, constfloat *vec3);
set the eigenvectors.
void setVector2(const float x, const float y, const floatz);
sets the second eigenvector to be.
void resetVector3(); sets the third eigenvector to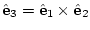
void setZero(); sets all the values to zero.
float value1() const; largest eigenvalueThe operators defined are:
float value2() const; second eigenvalue
float value3() const; smallest eigenvalue
float vector1(const int index) const; returns element ''index'' (01
2) of eigenvector 1
float vector2(const int index) const; returns element ''index'' (01
2) of eigenvector 2
float vector3(const int index) const; returns element ''index'' (01
2) of eigenvector 3
void print() const; sends the eigensystem to the standard output
bool isZero() const; check if eigensystem is zero (true if so)Functions are:
bool operator==(const eigen &e) const; compares all elements of the eigensystem, no tolerance
bool operator!=(const eigen &e) const; do.
bool operator< (const eigen &e) const; less then, uses measurement specified in measure()
bool operator> (const eigen &e) const; do.
bool operator<=(const eigen &e) const; do.
bool operator>=(const eigen &e) const; do.
void scale(const float factor); scale eigensystem by ''factor''Functions between two eigensystems:
void scale(const float x, const float y, const float z);
scale differently in each direction
void rotate(const float pitch, const float roll, const floatyaw);
Rotate eigensystem, the angles pitch, roll, and yaw
have to be provided in degrees
float det() const; determinant of the tensor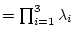
float relation_Lambda1_Lambda2() const; absolute value of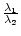
float relation_Lambda1_Lambda3() const; absolute value of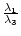
void eigen2tensor(tensor &t) const; converts an eigensystem into a tensor
int tensor2eigen(const tensor &t); converts a tensor in an eigensystem
negative return value is an error, 1 is success
measurement of how close diffusion tensor shape is to
float close_line() const; a line (equation 4.13)
float close_plane() const; a plane (equation 4.14)
float close_sphere() const; a sphere (equation 4.15)
anisotropy measure describing deviation
float anisotropy() const; from the spherical case(equation 4.17)
float cosinMaxEigen(const eigen &e) const;
angle between the vectors of the maximum eigenvalues
bool isSimilar(const eigen &e, const float tolerance, constint similarity) const;
compare 2 eigensystem, return 1 if identical, 0 else
bool hasSimilarShape(const eigen &e, const float tolerance)const
do.
bool hasSimilarSize(const eigen &e, const float tolerance) const;
do.
bool hasSimilarDirection(const eigen &e, const float tolerance) const;
do.
bool threshold(const float threshold=0.8) const;
return true if eigensystem above threshold
float readfloat(DOM_Node& node); read numerical valuesfrom node in xml file;Protected members that are only accessible to the derived classes are:
return -1 if no value present
read string from node in xml file;
return -1 if no value present
int readstring(DOM_Node& node, char* mystring);
read attribute as string from node in xml file;
return -1 if no value present
pre:mystring is the name of the attribute to get value
post: mystring is the value of the attribute;
int readattribute(DOM_Node& node, char* mystring);
void readfile(DOM_Node& node); read the location of the datafile from node in xml file (could be removed)
void readdata(DOM_Node& node); read the xml file and extract the data wanted;
void readmanualy(); read ALL values from console
void readmanualy(int& value); read value from console, exit if entered -1
void readmanualy(float& value); do.
void readmanualy(char* value); do.
void assert_values(); check if all values to write xml-header are present
if not, ask for values
int _dimx, _dimy, _dimz; the dimensions of the fieldAny new element added to the DTD should be added here.
float _deltax, _deltay, _deltaz; the distance betweenvoxels = voxel dimensions
char *_datafile; pointer to the data file
char *_orientation; orientation of the field: axial, coronal, saggital
char *_sourcefile; location of source to derive tensor file
char *_originalfile; location of the original data
baby specific data:
int _gestationalAge; Gestational age of the baby in weeks
int _mrNumber; Patient ID number
int _studyNumber; Study number
int _seriesNumber; Series number for the diffusion data
char * _date; Date of the scan
do everything needed to call readdata, should not needThe actual path to the data is generated in the derived class by appending the corresponding extension.
void read_XMLHeader( const char xmlFile[]);
any editing when changing DTD
void write_XMLHeader(const char xmlFile[]);
write a xml-header file at location xmlFile
set values
void setNumbers(const int age, const int patient_id, const int studyNr,
const int seriesNumber);
void setDate(const char* date);
void setOrig(const char* orig);
void setOrientation(const char orientation);
set the voxel dimensions for field
void setDelta(const float deltax, const float deltay, const float deltaz);
get the voxel dimensions for field
void getDelta(float& deltax, float& deltay, float& deltaz);
get the volume dimensions for field
void getDimension(int& dimx, int& dimy, int& dimz);
char * getPath(char * path); get the path to the data without any extension
No function to set the volume dimensions is provided, since this is either
already defined by the data set and therefore in the xml-header or a
new data set is created and then the corresponding derived class has
to set this values.
Finally three generic functions are included in this class:
check boundaries for offset and window-sizeThe last two functions read the content of the *.pic files generated by LSDI_recon.
and reset them if necessary.
used for display purposes:
Plane rotates the window depending on window-size
plane[3] = 2: Image is in xy-plane
= 1: Image is in xz-plane
= 0: Image is in yz-plane
in each case plane[0] and plane[1]
are the xy dimensions in the image coordinate system
plane[2] the number of images
void check_boundaries(int offset[3], int windowsize[3], int plane[4]=NULL) const;
short * read_pic(short *image,char path_file[], int offset=0);
unsigned short * read_pic(unsigned short *image, char path_file[], int offset=0);
|