| Visit DIX: German-Spanish-German dictionary | diccionario Alemán-Castellano-Alemán | Spanisch-Deutsch-Spanisch Wörterbuch |
This chapter discusses all the methods that have been used to align two tensorfields with a nonrigid registration. The following series of steps are used:
For simplicity in this section any field is called a three dimensional image ![]() with
values
with
values ![]() at position
at position ![]() . The values
. The values ![]() can
therefore be any of the types scalar, tensor or eigensystem. The stationary
image is
can
therefore be any of the types scalar, tensor or eigensystem. The stationary
image is ![]() and the moving
and the moving ![]() . The transformation from
. The transformation from ![]() to
to
![]() is denoted
is denoted ![]() :
:
| (7.2) |
| (7.3) |
The displacement field is
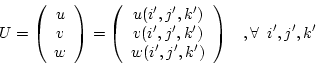 |
(7.4) |
A point ![]() at position
at position ![]() in
in ![]() in the stationary image has a
neighborhood
in the stationary image has a
neighborhood
![]() which is a small window of size
which is a small window of size
![]() . A neighborhood of a point
. A neighborhood of a point ![]() at position
at position
![]() in
in ![]() of
size
of
size
![]() is
referred to as
is
referred to as
![]() .
These notation conventions are summarized in Figure
7.1. Furthermore a collection of n points
.
These notation conventions are summarized in Figure
7.1. Furthermore a collection of n points
![]() with
with
![]() is denoted
is denoted
![]() .
.
Every point ![]() for which
for which ![]() is known can be displaced to a new
location
is known can be displaced to a new
location ![]() . If
. If ![]() is known for every point
is known for every point ![]() the
whole image
the
whole image ![]() can be displaced. The resulting image
can be displaced. The resulting image ![]() though
will have points
though
will have points ![]() which have not been assigned a value from
which have not been assigned a value from
![]() . That is, the function
. That is, the function ![]() is not directly invertible, since
is not directly invertible, since ![]() is not necessarily onto and not necessarily one to one.
is not necessarily onto and not necessarily one to one.
As a result an image ![]() is expected that has values
everywhere. Instead of interpolating the points
is expected that has values
everywhere. Instead of interpolating the points ![]() to get missing
values for
to get missing
values for ![]() the matching procedure can be inverted to get a
function
the matching procedure can be inverted to get a
function ![]() . This second approach is used here. Thus points are
selected in the stationary image
. This second approach is used here. Thus points are
selected in the stationary image ![]() . When
the corresponding points have been found and the displacement field
interpolated this leads to a displacement
. When
the corresponding points have been found and the displacement field
interpolated this leads to a displacement ![]() approximated by
approximated by ![]() from the stationary to the
moving image. Now for every position in a resulting image
from the stationary to the
moving image. Now for every position in a resulting image ![]() the
position where the values came from is known, and the transform is
onto, so that
the
position where the values came from is known, and the transform is
onto, so that
![]() is empty.
is empty.
A point on a line for example cannot be unambiguously matched to a point on a line in the second image if no additional information is provided. A corner instead has a unique counterpart in both images and can therefore be matched uniquely, of course only if the corner is present in both images. A measure of cornerness tells how much structure there is around a certain point.
In the scalar case the
derivatives of the image ![]() are computed and the outer product of the
resulting vector is built, which is by definition the correlation matrix
are computed and the outer product of the
resulting vector is built, which is by definition the correlation matrix ![]() . With the derivatives
. With the derivatives
| (7.6) |
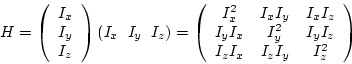 |
(7.7) |
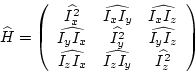 |
(7.8) |
![]() can be seen as a tensor of order 2. The eigenvalues can be
illustrated by an ellipsoid. The rounder the ellipsoid the bigger the
cornerness, i.e. the measure close-sphere in Equation
4.15 is a measure for cornerness in this case.
can be seen as a tensor of order 2. The eigenvalues can be
illustrated by an ellipsoid. The rounder the ellipsoid the bigger the
cornerness, i.e. the measure close-sphere in Equation
4.15 is a measure for cornerness in this case.
For any symmetric matrix the trace is the sum of the eigenvalues
| (7.9) |
| (7.10) |
Ruiz and co-workers [15] propose to use points above a threshold
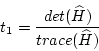 |
(7.11) |
Here a modified version is used, where the two thresholds are
incorporated in one formula. The structure ![]() is then defined as
is then defined as
Using the expectance causes blurring of the point-selection since high
values are diffused. The trace of ![]() has a non-zero value without
averaging and is an edge detector [1], as
has a non-zero value without
averaging and is an edge detector [1], as
There are now several options to improve the point-selection.
Before the process is started an anisotropic filter can be applied to
enhance edges. After using a point-selection as described the result can
be masked with the trace of ![]() (not
(not ![]() !) to undo the blurring
effect of the expectance computation. Finally the local maxima of a
region of the function 7.13 can be selected to get single voxel positions.
!) to undo the blurring
effect of the expectance computation. Finally the local maxima of a
region of the function 7.13 can be selected to get single voxel positions.
In the case of tensorfields the points should be extracted from a
structure with six independent values. For each independent component
of the tensor a matrix
![]() can be computed and the sum over the components of
these matrix used as a
final matrix
can be computed and the sum over the components of
these matrix used as a
final matrix ![]()
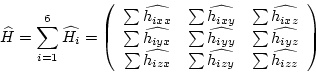 |
(7.15) |
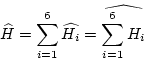 |
(7.16) |
The parameters for the point-selection in the program
nonrigidreg are described in Table A.3 in page ![[*]](crossref.gif) .
.
For each selected point ![]() in
in ![]() a neighborhood
a neighborhood
![]() is
selected. At the same position in the moving image
is
selected. At the same position in the moving image ![]() a search window
a search window
![]() is
selected, i.e. an area where the corresponding point is assumed to
lie. For each point
is
selected, i.e. an area where the corresponding point is assumed to
lie. For each point
![]() in this search-window a
neighborhood
in this search-window a
neighborhood
![]() is compared to the neighborhood
is compared to the neighborhood
![]() and a value measuring the result of the comparison
assigned to
and a value measuring the result of the comparison
assigned to ![]() . Two different measurements of similarity for scalar data are
implemented.
. Two different measurements of similarity for scalar data are
implemented.
Instead of simply selecting the best matching value the resulting
vectors NCC(![]() ) respectively LSE(
) respectively LSE(![]() ) are sorted so that NCC(
) are sorted so that NCC(![]() ),
LSE(
),
LSE(![]() ) are the best and NCC(
) are the best and NCC(![]() ), LSE(
), LSE(![]() )
the worst matches. Before accepting a match the sorted vectors can be
analyzed. If the best and worst match are too close to each other, i.e. NCC(
)
the worst matches. Before accepting a match the sorted vectors can be
analyzed. If the best and worst match are too close to each other, i.e. NCC(![]() )
) ![]() NCC(
NCC(![]() ),
there is not enough structure in the search-window
),
there is not enough structure in the search-window
![]() so that the match should be ignored. Furthermore if there are several
equally or almost equally good matches the displacement field in the
neighborhood should be considered and matching should additionally be
based on smoothness of the over-all displacement field. Only the first
approach has been implemented.
so that the match should be ignored. Furthermore if there are several
equally or almost equally good matches the displacement field in the
neighborhood should be considered and matching should additionally be
based on smoothness of the over-all displacement field. Only the first
approach has been implemented.
In a second step, when for all points
![]() a
correspondence has been found, the displacements are checked.
Overlapping displacements, i.e. displacement vectors that cross each
other are eliminated since the tissue of a subject should not be
folded.
Whenever two displacements cross each other the shorter
displacement is kept and the larger removed.
a
correspondence has been found, the displacements are checked.
Overlapping displacements, i.e. displacement vectors that cross each
other are eliminated since the tissue of a subject should not be
folded.
Whenever two displacements cross each other the shorter
displacement is kept and the larger removed.
In order to obtain a displacement field for the whole volume, the sparse displacements obtained at single locations have to be interpolated. This section describes the selected interpolation method. It is assumed that the sparse displacement field has an unknown underlying random field. References [35] and [38] give a good introduction and a simple example on how to use Kriging.
Kriging interpolation originates from geostatistics and is known to be the best linear unbiased estimator because it is theoretically capable of minimizing the estimation error variance while being a completely unbiased estimation procedure [34].
Kriging is a modified linear regression technique that estimates a
value at a point by assuming that the value is spatially related to
the known values in a neighborhood near that point. Kriging computes
the value for the unknown data point using a weighted linear sum of
known data values. The weights are chosen to minimize the estimation
error variance and to maintain unbiasedness in the sampling. Unlike
other techniques for scalar values, Kriging bases its estimates upon a
dynamic, not static, neighborhood point configuration and treats those
points as regionalized variables instead of random
variables. Regionalized variables assume the existence of regions of
influence in the data. In Kriging each region is analyzed to determine
the correlation or interdependence among the data in the
region and this is encoded through a function called a variogram. For
the unknown value ![]() at the position
at the position ![]() within the
neighborhood of known points
within the
neighborhood of known points ![]() with known values
with known values ![]() the basic Kriging equation is:
the basic Kriging equation is:
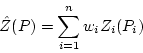 |
(7.19) |
![]() is the actual value at a point
is the actual value at a point ![]() , and
, and ![]() is the number of known
points used to compute
is the number of known
points used to compute ![]() . The
. The ![]() 's are the regionalized
variables and the
's are the regionalized
variables and the ![]() 's the weights. Unlike other techniques which
also use a weighted sums, in Kriging the weights are not selected
based solely upon the distance between sampled and unsampled points. Kriging
does not assume that the variability of the data is linear
[33].
's the weights. Unlike other techniques which
also use a weighted sums, in Kriging the weights are not selected
based solely upon the distance between sampled and unsampled points. Kriging
does not assume that the variability of the data is linear
[33].
Optimal weights are determined by enforcing the error expectation in the estimate be zero
| (7.20) |
and the error variance be minimal
| (7.21) |
where ![]() is the expected value or mean and
is the expected value or mean and
![]() the
mean-square-error of the dissimilarity between the two variables
the
mean-square-error of the dissimilarity between the two variables
![]() and
and ![]() .
These two conditions make
.
These two conditions make ![]() the best linear unbiased
estimator and are the base equations to derive the Kriging system of
equations. Furthermore the unbiasedness implies that the weights must
sum up to one:
the best linear unbiased
estimator and are the base equations to derive the Kriging system of
equations. Furthermore the unbiasedness implies that the weights must
sum up to one:
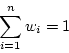 |
(7.22) |
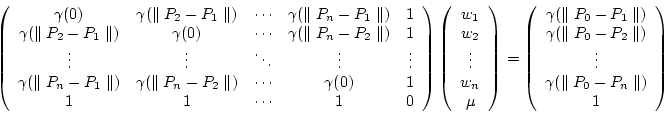 |
(7.23) |
In addition, there may be a discontinuity at the origin, the so-called
nugget effect. This means that the fitted model does not pass
through the origin, but intersects the y-axis at a positive value of
![]() , which is
, which is ![]() . This quantity is an estimate of
. This quantity is an estimate of ![]() , the
residual, that represents spatially uncorrelated
noise associated with any value of a random variable
, the
residual, that represents spatially uncorrelated
noise associated with any value of a random variable ![]() at
at ![]() .
This terminology is illustrated in Figure 7.3.
.
This terminology is illustrated in Figure 7.3.
The relationship between the variogram and the covariance is given by [36]
Table 7.1 is a collection of different
parametric correlation functions. Figure 7.4 shows the
semivariogram (Equation 7.24) for the functions in Table
7.1 with ![]() and
and ![]() .
.
In most cases the variogram is unknown and is approximated by a process called structural analysis. As there is no prior information on the resulting displacement field this approach is not applicable. Instead different variogram models are implemented so that the best model can be empirically determined by comparing the results of the registration process.
An example interpolation is shown where a synthetic example is randomly sampled at 10% and interpolated using the linear variogram model and two different neighborhoods (see Figure 7.5).
|
Three different local transformations are presented and discussed
here. The first is proposed in reference [15].
![]() is the tensor in image
is the tensor in image ![]() that is displaced by
that is displaced by ![]() to a new
position
to a new
position ![]() . The transformation is
. The transformation is ![]() . Then the local
transformation is applied on
. Then the local
transformation is applied on ![]() to get
the final tensor
to get
the final tensor ![]() at the position
at the position ![]() .
.
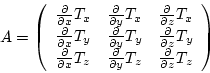 |
(7.25) |
| (7.26) |
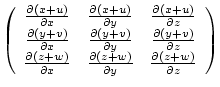 |
|||
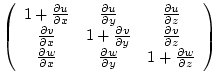 |
(7.27) |
![[*]](crossref.gif) ). The applied displacement field is
shown in Figure 7.6.
). The applied displacement field is
shown in Figure 7.6.
As can be seen, the shape of some tensors has been changed quite a bit.
| (7.31) |
Again, as was done for the rigid transformation, it is argued here to use this transformation when nonrigidly aligning the tensor images. When aligning two different subjects the structures should not be changed. In the scalar case gray-values are displaced to the new position but their value is not changed. Equivalently in the case of tensors, a local rotation is part of the transformation but changing the shape is not obvious and meaningful in all cases. The meaning of any local change would have to be studied for each application separately and even separately for each tissue, so that it is safest to not apply any strain. Also, if the point-extraction and the matching part of the nonrigid registration are based on measurements of the tensors that depend on the shape, changing the shape after the transformation leads to a change of the similarity. This can make the chosen displacement appear wrong after the local transformation.
![\includegraphics[width = 0.8\textwidth]{images/test_6_random_cut.eps}](img332.gif)
|
![\fbox{\includegraphics[width = 0.8\textwidth]{images/test_6_random_2.ps}}](img333.gif)
|
![\fbox{\includegraphics[width = 0.8\textwidth]{images/test_6_random_1.ps}}](img334.gif)
|
![\fbox{\includegraphics[width = 0.8\textwidth]{images/test_6_random_3.ps}}](img335.gif)
|
The implementation allows to choose between any of the described local
transformations (see Table A.3).
The size of the search-window is adapted to the current scale. For the lowest scale it is the specified search-window, divided by the number of scales times the scale-factor. When moving towards higher levels the search-window is a bit larger than twice the scale-factor, since any larger displacement should have been found in the lower levels.
Measuring the results, i.e. the improvement from
| (7.32) |
| (7.33) |
For the binary test data measuring the improvement is trivial and
boils down to be the number of voxel positions that have different
values in each image. The same principle can be used when matching
segmented data. Here the number of positions with different classes in each image is
a meaningful measure for dissimilarity. In grayscale data counting
positions where the graylevels are different is not very suggestive
since this will be the case almost everywhere in the body. Of course
the number should decrease as the border of the body should be better
aligned after the registration, but this will be a small
percentage. The total distance
| (7.34) |
| (7.35) |
When registering medical data the goal is to align the structures of
one data set to the other data set. Ideally the structures in both data
sets ![]() and
and ![]() are known, e.g. by segmenting the data. The
displaced structures can then be compared.
are known, e.g. by segmenting the data. The
displaced structures can then be compared.
Different tests are documented in Table 7.2. The parameters used are explained in Table A.3 in Appendix A.6. The process for the case of the chessboard is illustrated in Figure 7.11 and for the segmented baby brain in Figure 7.12.
|
|